 Figure 1.
Figure 1.Xerox PARC, 3333 Coyote Hill Road, Palo Alto, CA 94304. merkle@xerox.com
The URL for this document is: http://www.zyvex.com/nanotech/nanotechAndMedicine.html
This article has been published in Advances in Anti-Aging Medicine, Vol. I, edited by Dr. Ronald M. Klatz, Liebert press, 1996, pages 277-286. The material was first presented at the The 2nd annual conference on anti-aging medicine & biomedical technology for the year 2010, December 4-6 1994, Las Vegas Nevada.
This electronic article might differ in some respects from the published version.
Future advances in medical technology are usually of only academic interest to the patient of today. There is, though, a way to give today's patient access to future medical technology: cryonics. Though still controversial, it has greater potential to save lives than any other method that we can use today. A brief introduction to this subject with links to further reading is provided at http://www.ralphmerkle.com/cryo.
Nanomedicine, a new book series being written by Robert Freitas, covers the wide range of medical applications of nanotechnology in technical depth.
Nanotechnology, "the manufacturing technology of the 21st century," should let us economically build a broad range of complex molecular machines (including, not incidentally, molecular computers). It will let us build fleets of computer controlled molecular tools much smaller than a human cell and built with the accuracy and precision of drug molecules. Such tools will let medicine, for the first time, intervene in a sophisticated and controlled way at the cellular and molecular level. They could remove obstructions in the circulatory system, kill cancer cells, or take over the function of subcellular organelles. Just as today we have the artifical heart, so in the future we could have the artificial mitochondrion.
Equally dramatic, nanotechnology will give us new instruments to examine tissue in unprecedented detail. Sensors smaller than a cell would give us an inside and exquisitely precise look at ongoing function. Tissue that was either chemically fixed or flash frozen could be analyzed literally down to the molecular level, giving a completely detailed "snapshot" of cellular, subcellular and molecular activities.
Design concepts for general purpose self replicating manufacturing systems have been discussed for many years [REF10, REF27, REF28], and their utility in manufacturing has been emphasized recently [REF04, REF05, REF06, REF18, REF30]. These proposals draw on a body of work started by von Neumann[REF27]. A wide range of methods have been considered[REF10, particularly pages 190 et sequitur "Theoretical Background"]. The von Neumann architecture for a self replicating system is the ancestral and archetypal proposal[REF24, REF27].
 Figure 1.
Figure 1.
Von Neumann worked out the details for a constructor that worked in a theoretical two-dimensional cellular automata world (parts of his proposal have since been modeled computationally[REF24]). The constructor had an arm which it could move about and which could be used to change the state of the cell at the tip of the arm. By progressively sweeping the arm back and forth and changing the state of the cell at the tip, it was possible to create "objects" consisting of regions of the two-dimensional cellular automata world which were fully specified by the program that controlled the constructor.
While this solution demonstrates the theoretical validity of the idea, von Neumann's kinematic constructor (which was not worked out in such detail) has had perhaps a greater influence, for it is a model of general manufacturing which can more easily be adapted to the three-dimensional world in which we live. The kinematic constructor was a robotic arm which moved in three-space and which grasped parts from a sea of parts around it. These parts were then assembled into another kinematic constructor and its associated control computer.
An important point to notice is that self replication, while important, is not by itself an objective. A device able to make copies of itself but unable to make anything else would not be very valuable. Von Neumann's proposals centered around the combination of a universal constructor, which could make anything it was directed to make, and a universal computer, which could compute anything it was directed to compute. It is this ability to make any one of a broad range of structures under flexible programmatic control that is of value. The ability of the device to make copies of itself is simply a means to achieve low cost, rather than an end in itself.
Drexler's assembler follows the von Neumann kinematic architecture, but is specialized for dealing with systems made of atoms. The essential components in Drexler's assembler are shown in figure 2. The emphasis here (in contrast to von Neumann's proposal) is on small size. The computer and constructor both shrink to the molecular scale, while the constructor takes on additional detail consistent with the desire to manipulate molecular structures with atomic precision. The molecular constructor has two major subsystems: (1) a positional capability and (2) the tip chemistry.
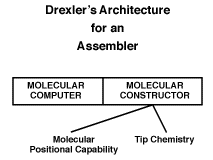 Figure 2.
Figure 2.
The positional capability might be provided by one or more small robotic arms, or alternatively might be provided by any one of a wide range of devices that provide positional control[REF09, REF15, REF25]. The emphasis, though, is on a positional device that is very small in scale: perhaps 0.1 microns (100 nanometers) or so in size.
The tip chemistry is logically similar to the ability of the von Neumann universal constructor to alter the state of a cell at the tip of the arm, but now the change in "state" corresponds to a change in molecular structure. That is, we must specify a set of well defined chemical reactions that take place at the tip of the arm, and this set must be sufficient to allow the synthesis of the structures of interest.
It is worth noting that current methods in computational chemistry are sufficient to model the kinds of structures that will appear in a broad class of molecular machines, including all of the structures and reactions needed for some assemblers[REF16, REF20, REF21, REF22]
Drexler's proposal for molecular mechanical logic [REF06] is the most compact and, from the system point of view, the best worked out. The logic elements ("locks," roughly the equivalent of a single transistor) need occupy a volume of only a few cubic nanometers. Even including system overhead (power, connections, etc). the volume per element should still be less than 100 cubic nanometers. A 10,000 element logic system (enough to hold a small processor) would occupy a cube no more than 100 nanometers on a side. That is, a volume only slightly larger than 0.001 cubic microns would be sufficient to hold a small computer. This compares favorably with the volume of a typical cell (thousands of cubic microns) and is even substantially smaller than subcellular organelles. Operating continuously at a gigahertz such a computer would use less than 10^-9 watts. By comparison, the human body uses about 100 watts at rest and more during exercise. Slower operation and the use of reversible logic would reduce power consumption, quite possibly dramatically[REF19, REF31].
A variety of molecular sensors and actuators would also fit in such a volume. A molecular "robotic arm" less than 100 nanometers long should be quite feasible, as well as molecular binding sites 10 nanometers in size or less.
By contrast, a single red blood cell is about 8 microns in diameter (over 80 times larger in linear dimensions than our 100 nanometer processor). Devices of the size range suggested above (~0.1 microns) would easily fit in the circulatory system and would even be able to enter individual cells.
Given such molecular tools, we could design a small device able to identify and kill cancer cells. The device would have a small computer, several binding sites to determine the concentration of specific molecules, and a supply of some poison which could be selectively released and was able to kill a cell identified as cancerous.
The device would circulate freely throughout the body, and would periodically sample its environment by determining whether the binding sites were or were not occupied. Occupancy statistics would allow determination of concentration. Today's monoclonal antibodies are able to bind to only a single type of protein or other antigen, and have not proven effective against most cancers. The cancer killing device suggested here could incorporate a dozen different binding sites and so could monitor the concentrations of a dozen different types of molecules. The computer could determine if the profile of concentrations fit a pre-programmed "cancerous" profile and would, when a cancerous profile was encountered, release the poison.
Beyond being able to determine the concentrations of different compounds, the cancer killer could also determine local pressure. A pressure sensor little more than 10 nanometers on a side would be sufficient to detect pressure changes of less than 0.1 atmospheres (a little over a pound per square inch. See, for example, the discussion on page 472 et sequitur of Nanosystems[REF06] for the kind of analysis involved. One atmosphere is ~10^5 Pascals, so PV in this case would be (0.1 x 10^5 ) x (10^-8)^3 or 10^4 x 10^ -24 or 10^-20 joules. Multiple samples would be required to achieve reliable operation, as kT is ~4 x 10^-21 joules at body temperature. Linear increases in sensor volume would produce exponential increases in immunity to thermal noise and linear improvements in pressure sensitivity if that were to prove useful. Doubling the linear dimensions of the sensor would produce an eight-fold increase in both volume and pressure sensitivity).
As acoustic signals in the megahertz range are commonly employed in diagnostics (ultrasound imaging of pregnant women, for example), the ability to detect such signals would permit the cancer killer to safely receive broadcast instructions. By using several macroscopic acoustic signal sources, the cancer killer could determine its location within the body much as a radio receiver on earth can use the transmissions from several satellites to determine its position (as in the widely used GPS system). Megahertz transmission frequencies would also permit multiple samples of the pressure to be taken from the pressure sensor, as the CPU would be operating at gigahertz frequencies.
The cancer killer could thus determine that it was located in (say) the big toe. If the objective was to kill a colon cancer, the cancer killer in the big toe would not release its poison. Very precise control over location of the cancer killer's activities could thus be achieved.
The cancer killer could readily be reprogrammed to attack different targets (and could, in fact, be reprogrammed via acoustic signals transmitted while it was in the body). This general architecture could provide a flexible method of destroying unwanted structures (bacterial infestations, etc).
6. An application: providing oxygen
This thickness, thin as it is, results in an applied stress on the diamond of well under 1% of its modulus -- from a purely structural point of view we should be able to use a very large "bucky ball," i.e., a sphere whose surface is a single layer of graphite. Perhaps the most complex issue involved in the selection of the material is the reaction of the body's immune system. While some suitable surface structure should exist which does not trigger a response by the immune system -- after all, there are many surfaces in the body that are not attacked -- the selection of a specific surface structure will require further research. To give a feeling for the range of possible surface structures, the hydrogenated diamond (111) surface could have a variety of "camouflauge" molecules covalently bound to its surface. A broad range of biological molecules could be anchored to the surface, either directly or via polymer tethers.
The Van der Waals' equation of state is (p+a/v^2) (v-b) = RT, where p is the pressure, v is the volume per mole, R is the universal gas constant, T is the temperature in Kelvins, and a and b are constants specific to the particular gas involved. For oxygen, a = 1.36 atm liter^2/mole^2 and b = 0.03186 liter/mole and R = 0.0820568 liter-atmospheres/mole-kelvin. A mole of oxygen at 1,000 atmospheres and at body temperature (310 Kelvins) occupies 0.048 liters, or about 21 moles/liter. A mole of oxygen at 1 atmosphere and 310 Kelvins occupies 25.4 liters, or about 0.04 moles/liter. This implies a compression of ~530 to 1. A resting human uses ~240 cc/minute[REF32] of oxygen, so a liter of oxygen compressed to 1,000 atmospheres should be sufficient to maintain metabolism for about 36 hours (a day and a half). It might be desirable to replace less than a liter of blood with our microspheres of compressed oxygen, but it should still be quite feasible to provide oxygen to tissue even when circulation is severely compromised for periods of at least many hours from a single infusion.
Controlled release of oxygen from the diamondoid sphere could be done using the selective transport method proposed by Drexler[REF06] and illustrated in figure 3. Figure 3 shows transport in the "wrong" direction (for this application), but simply reversing the direction of rotor motion would result in transport from inside the reservoir to the external fluid. By driving a rotor at the right speed, oxygen could be released from the internal reservoir into the external environment at the desired rate.
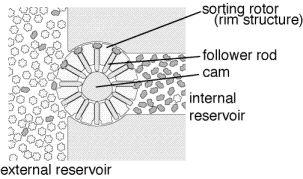 Figure 3.
Figure 3.
More sophisticated systems would release oxygen only when the measured external partial pressure of oxygen fell below a threshold level, and so could be used as an emergency reserve that would come into play only when normal circulation was (for some reason) interupted.
Full replacement of red blood cells would involve the design of devices able to absorb and compress oxygen when the partial pressure was above a high threshold (as in the lungs) while releasing it when the partial pressure was below a lower threshold (as in tissues using oxygen). In this case, selective transport of oxygen into an internal reservoir (by, for example, the method shown in Figure 3) would be required. If a single stage did not provide a sufficiently selective transport system, a multi-staged or cascaded system could be used. Compression of oxygen would presumably require a power system, perhaps taking energy from the combustion of glucose and oxygen (thus permitting free operation in tissue). Release of the compressed oxygen should allow recovery of a significant fraction of the energy used to compress it, so the total power consumed by such a device need not be great.
If the device were to simultaneously absorb carbon dioxide when it was present at high concentrations (in the tissue) and release it when it was at low concentrations (in the lungs), then it would also provide a method of removing one of the major products of metabolic activity. Calculations similar to those given above imply a human's oxygen intake and carbon dioxide output could both be handled for a period of about a day by about a liter of small spheres.
As oxygen is being absorbed by our artificial red blood cells in the lungs at the same time that carbon dioxide is being released, and oxygen is being released in the tissues when carbon dioxide is being absorbed, the energy needed to compress one gas can be provided by decompressing the other. The power system need only make up for losses caused by inefficiencies in this process. These losses could presumably be made small, thus allowing our artificial red blood cells to operate with little energy consumption.
By comparison, a liter of blood normally contains ~0.2 liters of oxygen[REF32, page 1722], while one liter of our spheres contained ~530 liters of oxygen (where "liter of oxygen" means, as is common in the literature on human oxygen consumption, one liter of the gas under standard conditions of temperature and pressure). Thus, our spheres are over 2,000 times more efficient per unit volume than blood; taking into account that blood is only about half occupied by red blood cells, our spheres are over 1,000 times more efficient than red blood cells.
Failure of a 0.1 micron sphere would result in creation of a bubble of oxygen less than 1 micron in diameter. Occasional failures could be tolerated. Given the extremely low defect rates projected for nanotechnology, such failures should be very infrequent.
While providing oxygen to healthy tissue should maintain metabolism, tissues already suffering from ischemic injury (tissue injury caused by loss of blood flow) might no longer be able to properly metabolize oxygen. In particular, the mitochondria will, at some point, fail. Increased oxygen levels in the presence of nonfunctional or partially functional mitochondria will be ineffective in restoring the tissue. However, more direct metabolic support could be provided. The direct release of ATP, coupled with selective release or absorption of critical metabolites (using the kind of selective transport system mentioned earlier), should be effective in restoring cellular function even when mitochondrial function had been compromised. The devices restoring metabolite levels, injected into the body, should be able to operate autonomously for many hours (depending on power requirements, the storage capacity of the device and the release and uptake rates required to maintain metabolite levels).
While levels of critical metabolites could be restored, other damage caused during the ischemic event would also have to be dealt with. In particular, there might have been significant free radical damage to various molecular structures within the cell, including its DNA. If damage was significant restoring metabolite levels would be insufficient, by itself, to restore the cell to a healthy state. Various options could be pursued at this point. If the cellular condition was deteriorating (unchecked by the normal homeostatic mechanisms, which presumably would cease to function when cellular energy levels fell below a critical value), some general method of slowing further deterioration would be desirable. Cooling of the tissue, or the injection of compounds that would slow or block deteriorative reactions would be desirable. As autonomous molecular machines with externally provided power could be used to restore function, maintaining function in the tissue itself would no longer be critical. Deliberately turning off the metabolism of the cell to prevent further damage would become a feasible option. Following some interval of reduced (or even absent) metabolic activity during which damage was repaired, tissue metabolism could be restarted again in a controlled fashion.
It is clear that this approach should be able to reverse substantially greater damage than can be dealt with today. A primary reason for this is that autonomous molecular machines using externally provided power would be able to continue operating even when the tissue itself was no longer functional. We would finally have an ability to heal injured cells, instead of simply helping injured cells to heal themselves.
Advances in medical technology necessarily depend on our understanding of living systems. With the kind of devices discussed earlier, we should be able to explore and analyze living systems in greater detail than ever before considered possible.
Autonomous molecular machines, operating in the human body, could monitor levels of different compounds and store that information in internal memory. They could determine both their location and the time. Thus, information could be gathered about changing conditions inside the body, and that information could be tied to both the location and the time of collection. Physical samples of small volumes (nano tissue samples) could likewise be taken.
These molecular machines could then be filtered out of the blood supply and the stored information (and samples) could be analyzed. This would provide a picture of activities within healthy or injured tissue. This new knowledge would give us new insights and new approaches to curing the sick and healing the injured.
More dramatically, it should be feasible to take "snapshots" of tissue samples and analyze the structure down to the molecular level. First, a small tissue sample could be either fixed or frozen. Chemical fixation can be used to rapidly block most tissue changes. Ultra fast freezing of small tissue samples is an effective method of halting essentially all chemical processes and diffusion of all molecules.
Once fixed or frozen, the tissue sample could be analyzed in a leisurely fashion. With nanotechnology (and indeed, to some extent with current STM and AFM technologies, though rather more expensively) it should be feasible to scan the tissue surface in molecular detail, and store that information in a computer. Once the surface had been scanned, it could be removed in a very selective and precise fashion, and scanned again. As an example, the use of a positionally controlled carbene has been proposed for use in the synthesis of complex diamondoid structures [REF06, REF21]. Such a positionally controlled carbene is highly reactive and, if positioned at an appropriate site on the surface of the tissue being analyzed, would readily react with a surface molecule. This surface molecule could then be removed. A wide variety of other "sticky" molecular tools could be brought up to the surface and allowed to react with surface molecules, which could then be removed, exposing the layers beneath.
The use of a positionally controlled carbene implies that the environment in which it is used must be inert. This requirement could be satisfied by analyzing the tissue sample at very low temperature (a few Kelvins) and in a very good vacuum. Under these conditions the tissue specimen would remain stable during even a protracted analysis process.
While this process can readily be envisioned for very small structures, nanotechnology should make massive parallelism feasible. That is, a single positional device could be used at a certain speed to provide information about a certain (rather small) volume of tissue in a reasonable time. Nanotechnology should permit the manufacture of a large number of small devices, each able to analyze a small volume. Given enough such devices operating in parallel, larger volumes could be analyzed and the information from many individual devices integrated to provide a coherent picture of the larger whole. Effective use of this option will require massive computational power -- which will also be made feasible with nanotechnology. Estimates of the computational power that should be provided by nanotechnology exceed 10^24 logic operations per second for a single desktop computer[REF06]. This amount of raw computational power should make control of a large number of parallel devices feasible, and should permit integration and analysis of the information so obtained.
In short, tissue samples could be "frozen" (either literally by ultrafast cooling or figuratively by chemical fixation) and the entire resulting tissue sample could be analyzed down to the level of individual molecules. The information so obtained could be processed by computers able to handle the flood of data produced. The resulting "snapshots" will provide us with an instantaneous look at metabolic and cellular activities across even relatively large volumes of tissue. Such an ability should revolutionize our understanding of the complex processes that take place in living systems. The possibility of truly revolutionary advances in our medical abilities has also created renewed interest in cryonics.
The abilities discussed here might well take years or decades to develop. It is quite natural to ask: "When might we see these systems actually used?" The scientifically correct answer is, of course, "We don't know." That said, it is worth noting that if progress in computer hardware continues as the trend lines of the last 50 years suggest, we should have some form of molecular manufacturing in the 2010 to 2020 time frame. After this, the medical applications will require some additional time to develop.
The remarkably steady trend lines in computer hardware, however, give a false sense that there is a "schedule" and that developments will spontaneously happen at their appointed time. This is incorrect. How long it will take to develop these systems depends very much on what we do. If focused efforts to develop molecular manufacturing and its medical applications are pursued, we will have such systems well within our lifetimes. If we make no special efforts the schedule will slip, possibly by a great deal.
As might be appreciated, developing these systems within our lifetimes would be advantageous for a variety of reasons.
REF04. Drexler, K.E. (1981) Molecular engineering: an approach to the development of general capabilities for molecular manipulation, Proc. National Academy of Sciences USA, 78, pp. 5275-8.
REF05. Drexler, K. E. (1986) Engines of creation, Doubleday.
REF06. Drexler, K.E., (1992) Nanosystems: molecular machinery, manufacturing, and computation , Wiley&Sons.
REF07. Eigler, D.M., and Schweizer, E.K. (1990) Positioning single atoms with a scanning tunnelling microscope, Nature 344, 524-526.
REF08. Feynman, R.P. (1960) There's plenty of room at the bottom, Caltech's Engineering and Science, February 1960, pages 22-36.
REF09. Fitzgerald, J.M. and Lewis, F.L. (1993) Evaluating the stewart platform for manufacturing, Robotics today, 6, pp. 1-3.
REF10. Freitas, R.A., and Gilbreath, W.P., (1980) Advanced automation for space missions, National Technical Information Service N83-15348.
REF15. Klafter, R.D., Chmielewski, T.A., and Negin M. (1989) Robotic engineering: an integrated approach, Prentice Hall.
REF16. Merkle, R.C. (1991) Computational nanotechnology, Nanotechnology , 2, pp. 134-141.
REF18. Merkle, R.C. (1992) Self replicating systems and molecular manufacturing, Journal of the British Interplanetary Society, 45, pp. 407-413.
REF19. Merkle, R.C. (1993) Reversible electronic logic using switches, Nanotechnology , 4, pp. 21-40.
REF31. Merkle, R.C. (1993) Two types of mechanical reversible logic, Nanotechnology 4 pages 114-131
REF20. Merkle, R.C. (1993) A proof about molecular bearings, Nanotechnology , 4 pp. 86-90.
REF21. Merkle, R.C. (1993) Molecular manufacturing: adding positional control to chemical synthesis, Chemical Design Automation News, 8, No. 9&10, pp. 1.
REF30. Merkle, R.C. (1994) Self replicating systems and low cost manufacturing, in Welland, M.E. and Gimzewski, J.K.(eds) The Ultimate Limits of Fabrication and Measurement, Kluwer Dordrecht.
REF32. Mountcastle, V.B (1980) Medical Physiology, Fourteenth Edition, Mosby.
REF22. Musgrave, C.B., Perry, J.K., Merkle, R.C., and Goddard, W.A. (1992) Theoretical studies of a hydrogen abstraction tool for nanotechnology, Nanotechnology , 2, pp. 187-195.
REF24. Signorini, J. (1989) How a SIMD machine can implement a complex cellular automaton? [sic] A case study: von Neumann's 29-state cellular automaton, in Proceedings Supercomputing `89, ACM Press.
REF25. Stewart, D (1965-66) A platform with six degrees of freedom, The Institution of Mechanical Engineers, Proceedings 1965-66, 180 Part 1, No. 15, pp. 371-386.
REF27. Von Neumann, J. and Burks, A.W. (1966) Theory of selfreproducing automata, University of Illinois Press.
REF28. Von Tiesenhausen, G., and Darbro, W.A. (1980) Self-replicating systems - a systems engineering approach, NASA technical memorandum TM-78304, Marshall Space Flight Center, Alabama.
 This
page is part of the
nanotechnology web site.
This
page is part of the
nanotechnology web site.